Improving interoperability in digital HER2 FISH enumeration a pilot evaluation



Background and Introduction:
Tissue matching between H&E or IHC slides and HER2 FISH specimens allow for specific targeting of tumor regions with the highest protein expression, optimizing the high-magnification scanning of the FISH slide. Traditionally, this workflow was performed on the same scanning platform in the FISH lab. The present evaluation aims to assess the feasibility of matching a brightfield image acquired on one scanning platform, with a FISH image acquired on a different system, thus enabling the use of the region of interest marked by the pathologist on the H&E or IHC image without requiring a second rescan.
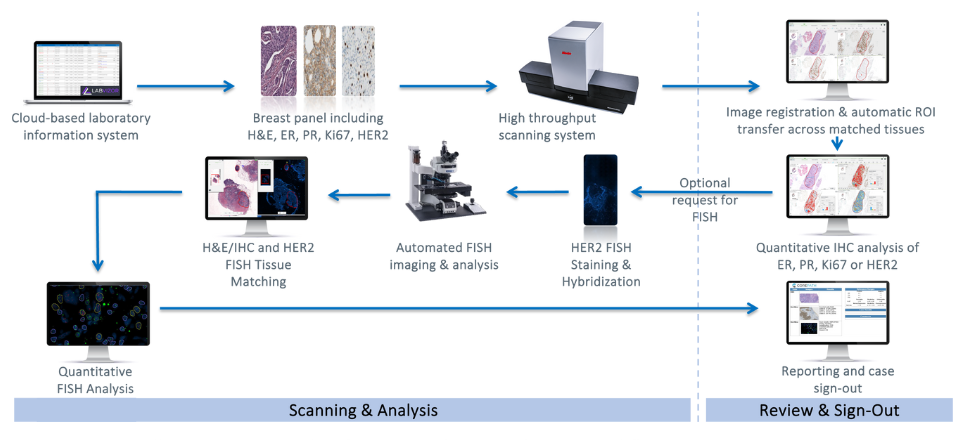
Figure 1: Illustrative example of integrated IHC and FISH workflows
Design and Method:
Core biopsy specimens from breast cancer patients were included in this evaluation. H&E and HER2 IHC slides were scanned with MoticEasyScan Infinity at 40X resolution (0.26um/px) and saved in SVS format. Certified pathologists examined slides using both conventional microscopy and digitalized imaging. FISH was requested in cases of equivocal reporting, and analysis was performed manually. The FISH slides were then scanned and analyzed using the PathFusion system (Applied Spectral Imaging). The brightfield images, acquired on the MoticEasyScan and marked by the pathologist when requesting FISH, were matched to the FISH images acquired on the ASI system in the FISH lab. Regions of interest were automatically transferred from the brightfield image to the FISH scan, and frames were selected in these marked areas for high-magnification scanning. Results of digital FISH enumeration were compared to manual results.
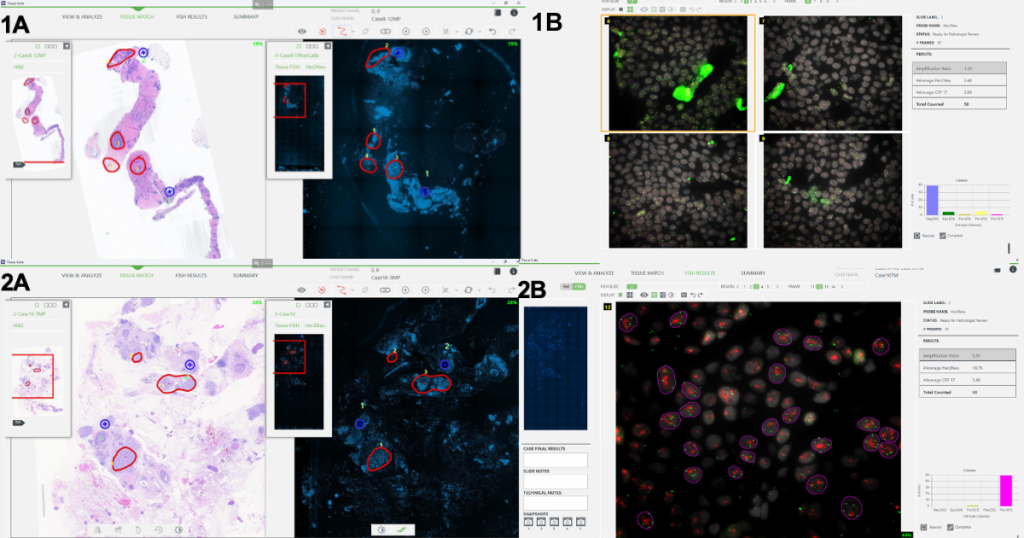
Figure 2: Representative examples of FISH negative case #8 (1) and FISH positive case #10 (2) featuring tissue matching between MoticEasyScan brightfield image and ASI FISH scan (A) and computer-aided FISH analysis results (B)
Case # | Manual HER2 FISH | Computer-aided HER2 FISH |
|---|---|---|
1 | 1.1 NEG | 1.1 NEG |
4 | 1.1 NEG | 1.1 NEG |
5 | 1.0 NEG | 1.3 NEG |
8 | 1.3 NEG | 1.2 NEG |
10 | 4.9 POS | 5.4 POS |
17 | 3.8 POS | 2.8 POS |
18 | 3.5 POS | Not available (faded signals) |
Table 3: Compared results of manual and computerized HER2/CEN17 FISH amplification ratio
Results:
Twenty biopsy specimens from 20 patients were included in this evaluation. Nineteen samples were diagnosed with invasive ductal or lobular carcinoma, and one with metaplastic carcinoma with chondroid differentiation. Among the 20 samples, 7 were diagnosed as HER2 IHC equivocal (2+). Manual HER2 FISH enumeration confirmed 3 as HER2 positive and 4 as HER2 negative. FISH specimens were then scanned in the FISH lab, and high-magnification frames were acquired in regions of interest marked by the pathologist on the brightfield image, eliminating the need to review the FISH slide under the microscope. Comparison to the manual process showed that digital FISH enumeration provided equivalent results for 6 slides. In one specimen, FISH signals were faded and unusable for digital enumeration.
Conclusion:
Interoperability in digital FISH enumeration allows the use of regions of interest marked by the pathologist on the H&E or IHC image when requesting FISH. This integrated workflow is envisioned to enhance both accuracy and efficiency in performing digital HER2 FISH enumeration.